Concept
- Problems
- Solutions
Engineering for nature
Biodiversity preservation has become a major challenge of the 21st century. The classical conservation and taxonomy approach no longer fit the emergency of the situation.
To fill the gap between new genomic conservation approaches and DNA sequencing technologies, GenoRobotics is engaging in:
- Plant biodiversity identification through the RaPid project
- Microorganisms identification in fresh water through the project CoWaS
- Plankton identification in sea water through the project JAWS in collaboration with Plankton Planet
- Interfacing the DNA extraction devices through interfacing tools
- Analysis of the sampled DNA with consensus sequence research
- Data storage and comparison through the database implementation
Plants
RaPID

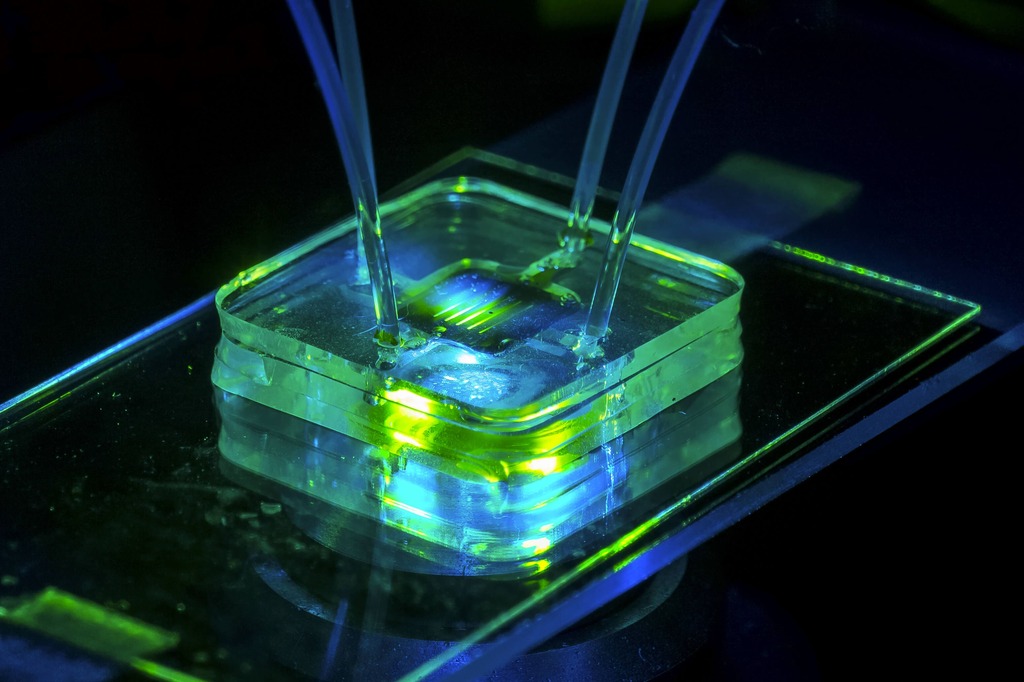
Forest represent a large variety of samples as trees, plants and mushrooms. These species provide many services to humans, but these are not free. Responsible management and knowledge of ecosystems is necessary to act appropriately.
The DNA is located deep in the cell nucleaus, itself protected by two protective layers: the cell wall and the cell membrane.
To extract DNA, we are developing a miniaturized tool based on hydrogels with our microneedles technology for field DNA sequencing allowing:
- A large scale of plant sampling
- Automated DNA extraction
- DNA quantification
Water
CoWaS & JAWS
Water as mountain lake river and ocean represent a new challenge to overcome. Access to these environments is not easy, but crucial because the water cycle is a key component of life on earth. The observation of aquatic biodiversity allows the monitoring of many environmental variables.
The goal is to accelerate the classification thanks to environmental DNA.
We are developing a new tool for remote DNA analysis of water samples. We are currently working on:
- Automation of water sampling
- Continuous DNA extraction
CoWaS is developped for fresh water use cases while JAWS is targeted toward salty sea water.


Raw data
Interfacing tools


Getting physically genomic information is one step (done with RaPId), having this information in the form of usable bytes in the computer is another.
We are developping desktop/web-based applications to enable user-friendly data transfers/upload/download from the sensors to our computers and database.
Coming soon.
Processed data
Database
Identification and sequencing requires huge amounts of data. A database is created to store and manage those data.
Many international database already exist such as CenBank. The goal is to design an interface that enables data exchange between these systems and ours.
Coming soon


Genome sequences
Consensus sequence
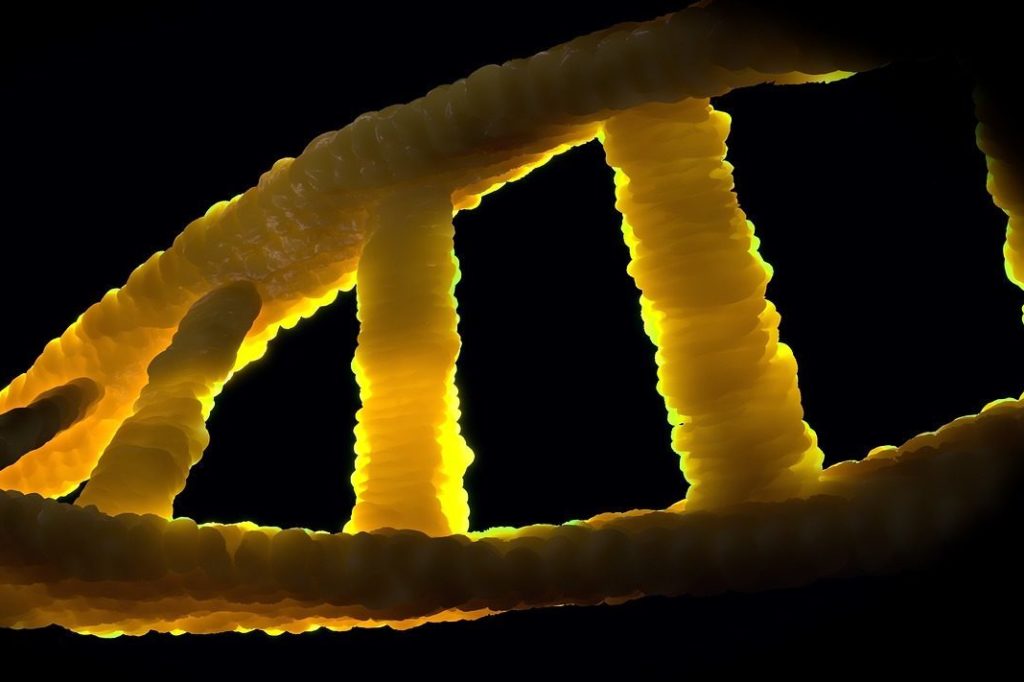
DNA, the code of life, is composed of sequences called genes, composing the genome of all species. The quantity of genes for one DNA strand is enormous. To identify a specie, the whole genome is not needed, also it would not be viable to store all this heavy data. Biologists use something called consensus sequences. Some short pieces of genes which are identified as unique for each specie. Isolating correctly such a sequence is the key to biodiversity identification.
Coming soon
F.A.Q.
Master the tech.
